Research Areas
The Klem Lab develops integrative computational models to understand and exploit enzyme function. We combine quantum mechanics (QM), molecular dynamics (MD), and machine learning to design biocatalysts, reveal mechanisms, and support real-world applications in medicine and sustainability.
1. Computational Tools for Enzyme Modeling
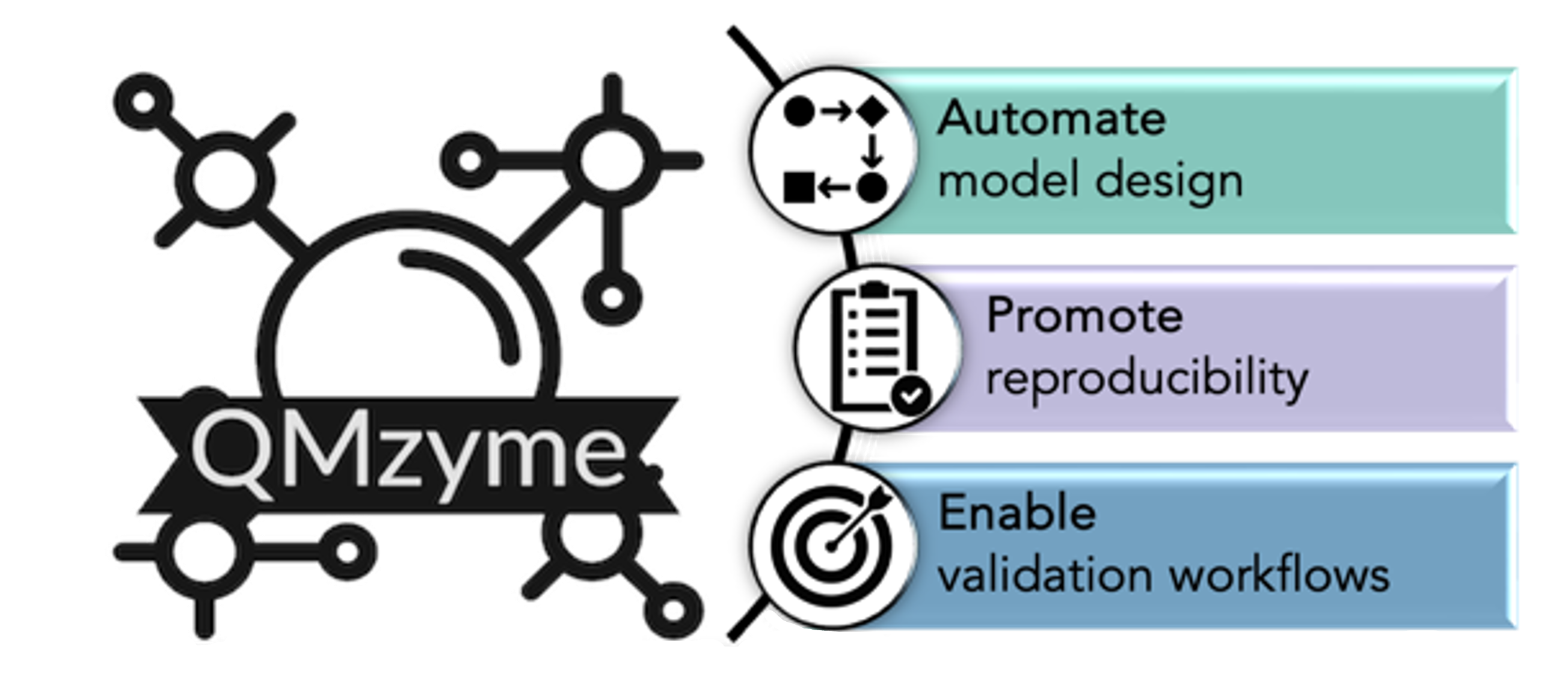
We are committed to building accessible, reproducible, and community-driven software tools to support the broader enzymology and computational chemistry communities. Our flagship platform, QMzyme, automates quantum mechanical based enzyme model generation and is actively expanding to include PyMOL plugins, high-throughput interfaces, and educational features. We also create standardized workflows and infrastructure that promote best practices and reproducibility in computational enzymology. We implement our developed multiscale workflows that unify high-fidelity quantum mechanics and molecular dynamics to investigate enzymatic mechanisms with particular interests in elucidating electric field effects. From understanding β-lactamase resistance to advancing the design of active site mimetics, our approach integrates electronic structure methods and conformational analysis to ensure a comprehensive understanding of nature’s complex catalysts. A key goal is to identify electrostatic and structural features that inform antibiotic development and de novo enzyme design.
2. Automation and Machine Learning for Biocatalyst Design
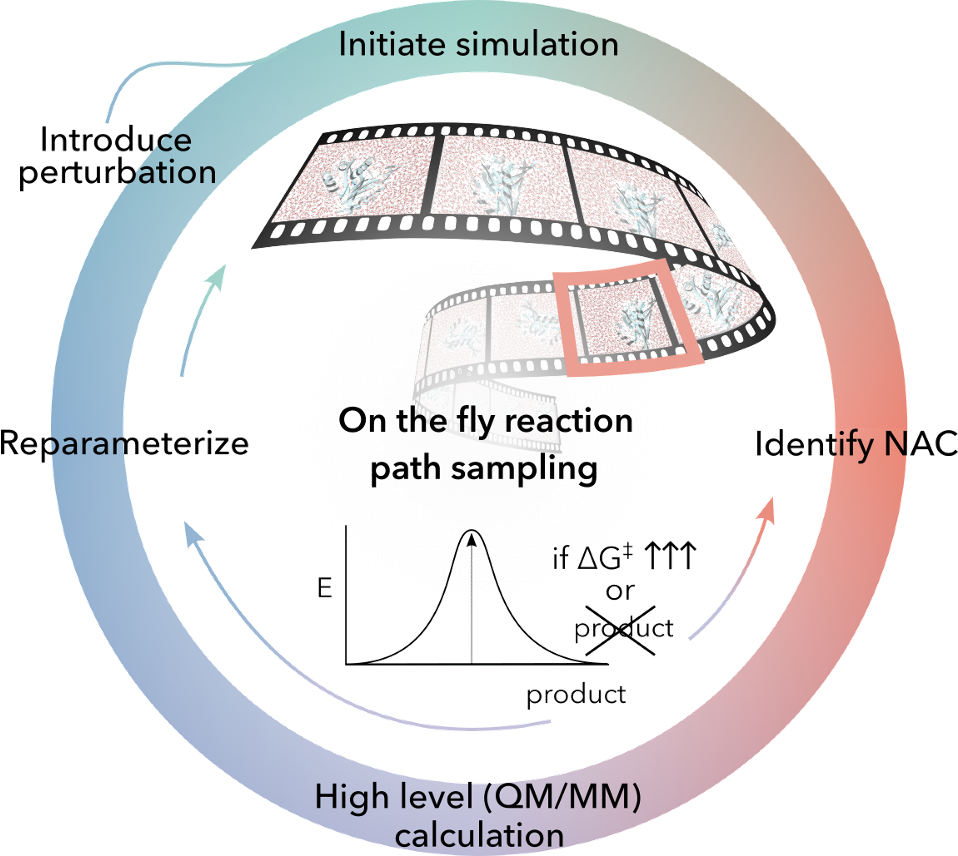
We streamline biocatalyst engineering by combining automated QM/MM workflows with machine learning models that predict and guide enzyme optimization. These integrated approaches accelerate discovery by reducing reliance on costly trial-and-error experiments. Current projects include mapping near-attack conformations, studying catalytic promiscuity, and integrating feedback from ML models with physics-based calculations.
3. Multienzyme Systems for Plastic Degradation
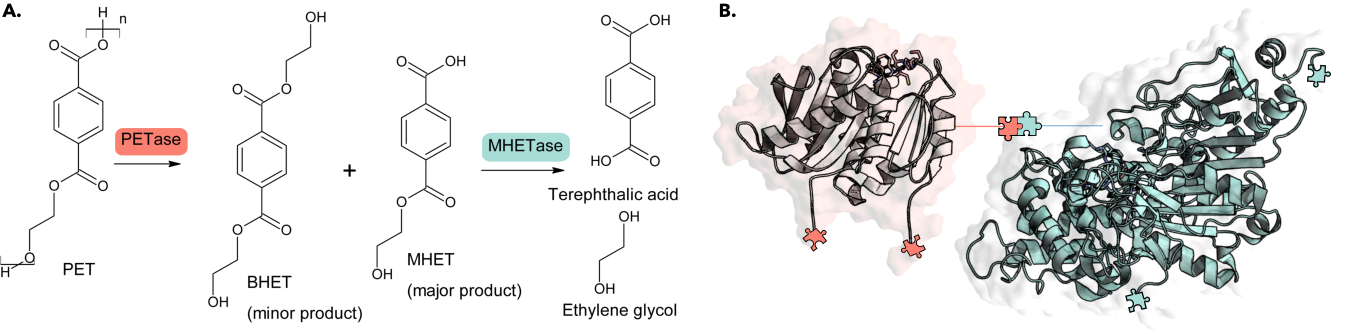
We apply our modeling expertise to tackle environmental plastic waste through enzyme engineering, focusing first on PET bio-depolymerization. Our goal is to improve plastic depolymerization through computational insights into mechanism, structure-function relationships, and data-driven mutagenesis prioritization.